Main Interface
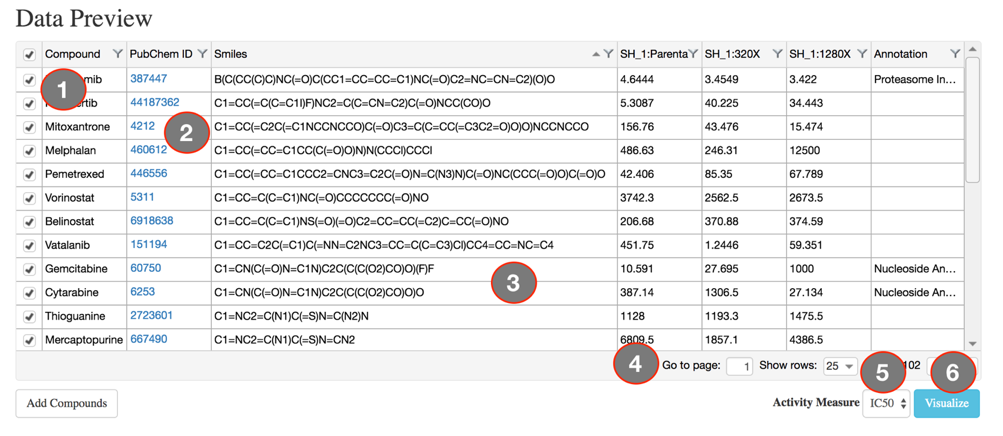
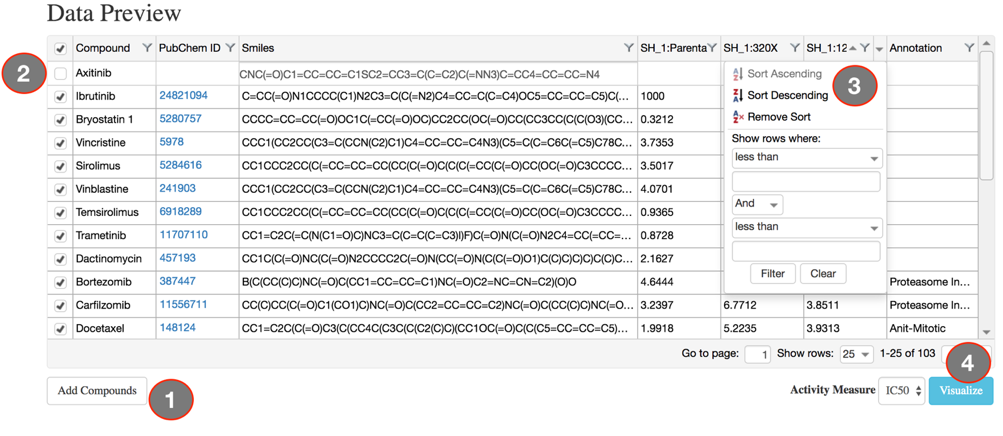
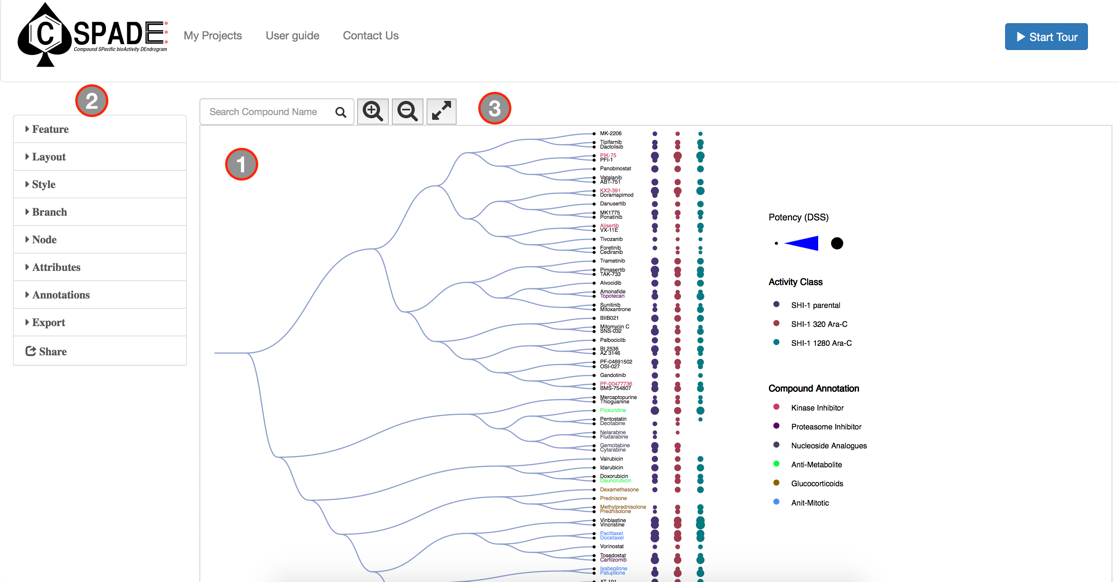
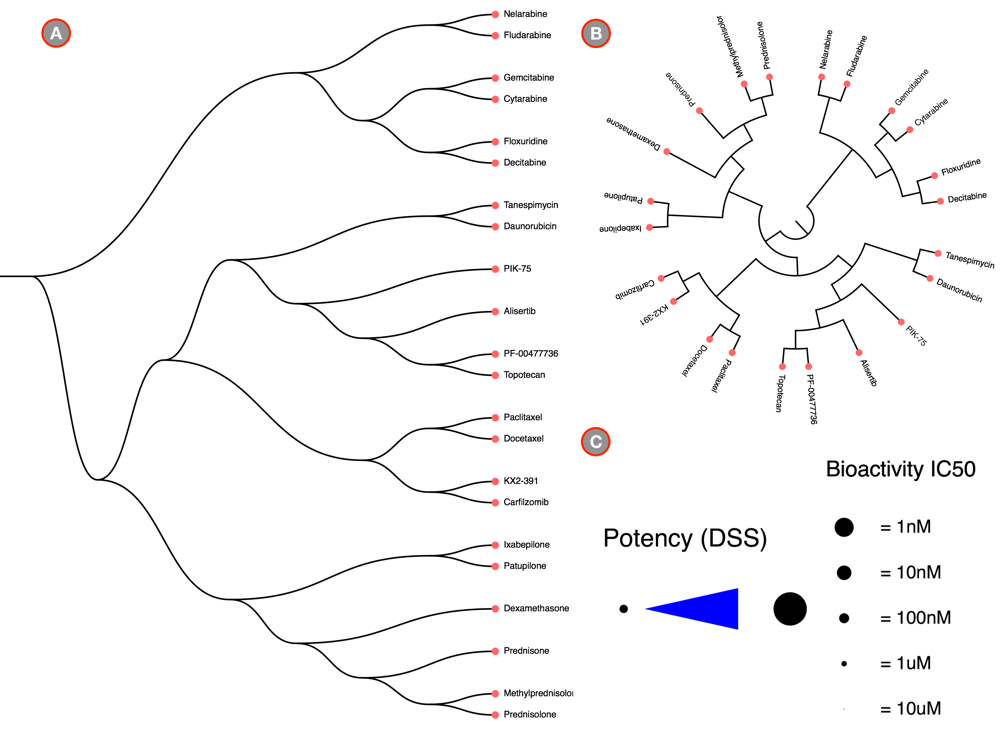
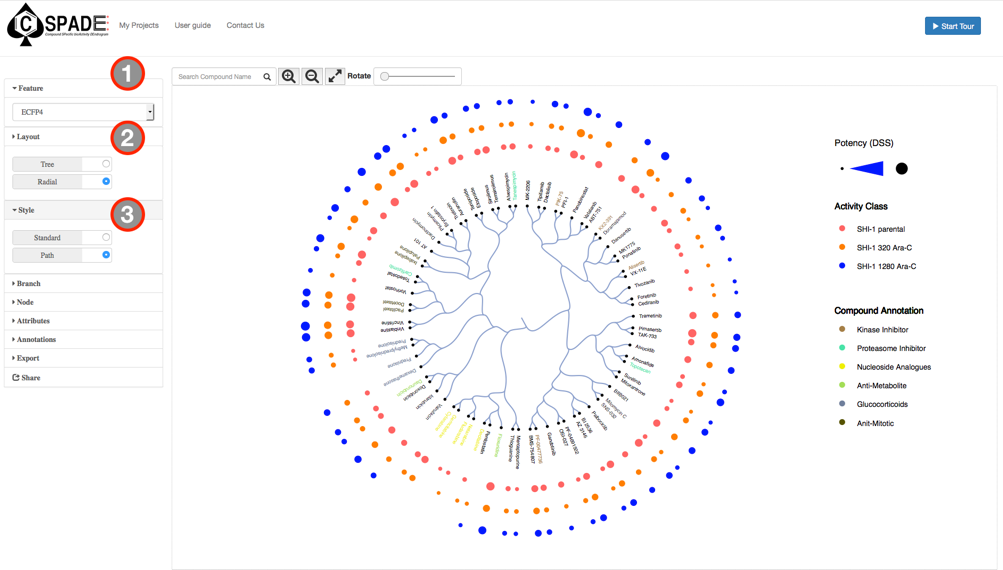
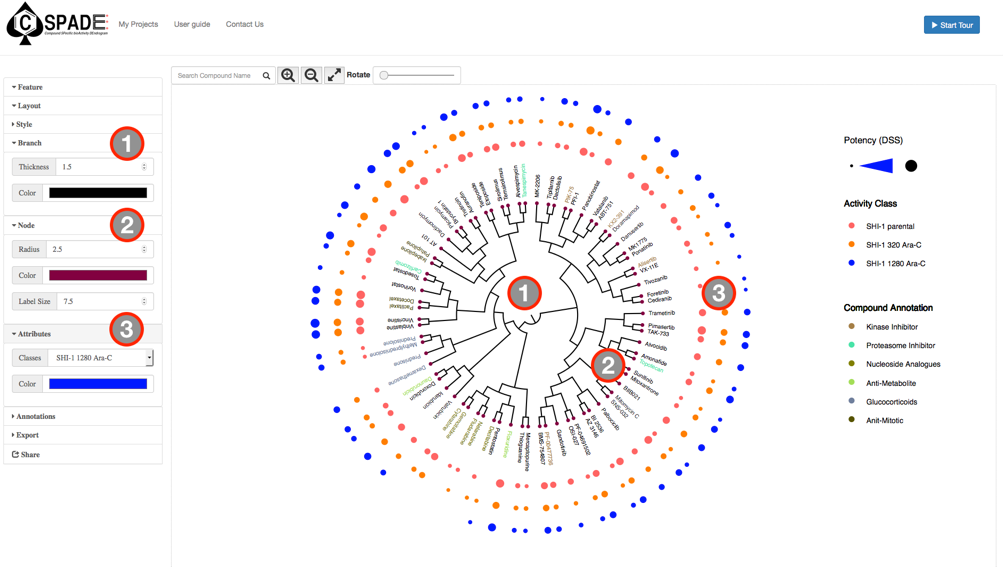
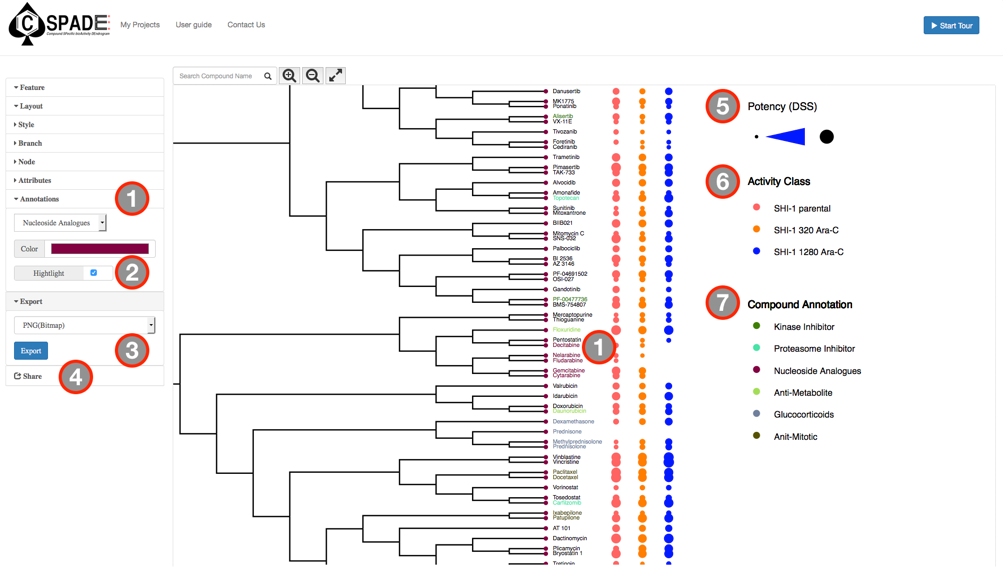
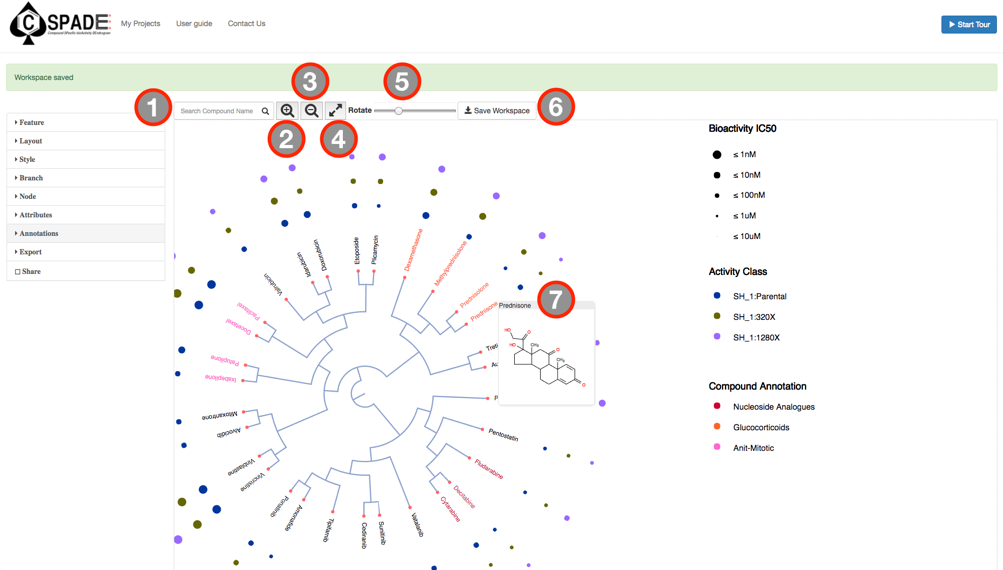
The main visual interface displays the compound similarity dendrogram, by default the compound similarity is estimated using the ECFP4 fingerprints, represented as Tree and the edges displayed as Path. The compounds that were selected in the Data Preview Page will be the nodes and the distance between two nodes is an estimate of their similarity. The compound names will be the labels of the node and the activity measure given in the input data are categorized into five potency classes using log-transformed IC50, EC50, Ki, Kd values (≤1nM, ≤10nM, ≤100nM, ≤1uM, ≤10uM); With the the summary measurements, such as AUC and DSS, the actual bioactivity values are used and represented as circular annotations in the dendrogram, the radius of which corresponds to the categorized bioactivity measure provided and is displayed in the Bioactivity legend and the color coding uniquely representing the various assays are shown in the Activity Classes legend. Annotation of the compounds if provided will be displayed with unique color and shown in the Compound Annotation legend.